MALDI-TOF MS techniques in microbial identification
Bioanalytics, Contamination, Microbiology

- MALDI-TOF mass spectrometry is a highly sensitive, cost-effective, and fast method for identification of bacterial, viral and fungal species. Its applications continuously expand but the most important one is rapid microorganism detection in pharmaceutical industry.
- The technique analyzes ribosomal proteins unique to specific microorganisms by creating a mass spectrum “fingerprint” compared against a database for identification. This allows for the identification of up to 96 isolates in just 1.5 hours.
- While highly effective, the technique faces challenges such as smaller library spectra for environmental strains and the need for improved databases and sample preparation methods. Despite these challenges, MALDI-TOF MS remains a promising tool for various microbiological applications due to its speed, precision, and cost-efficiency.
Background
Matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (MS) has been efficiently used for over 30 years. This revolutionary technique offers high sensitivity with short analysis time, robustness, cost-effectiveness, high throughput and ease of use compared to other analytical techniques.1 MALDI‑TOF MS has been widely implemented in microbiological laboratories for routine identification of various bacterial and fungal species. Its range of applications has been steadily growing, extending from rapid identification of organisms to laborious proteomic studies of bacterial physiology.2 The versatility and speed of analysis have led to the development of numerous MALDI‑TOF methods involving bacteria such as detection of recombinant proteins, analysis of bacterial DNA and RNA, detection of virulence markers and, already mentioned, rapid characterization of bacteria at the genus, species, and strain level.3
Performance and application of MALDI-TOF mass spectrometry method in microbiology laboratories
The MALDI-TOF MS method is based on the analysis of ribosomal proteins, which are abundant in microorganisms but unique to specific family, genus and species, or even strain.
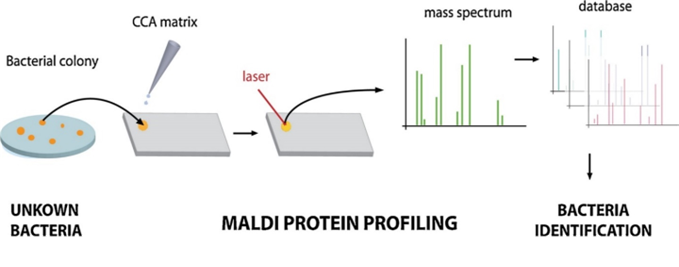
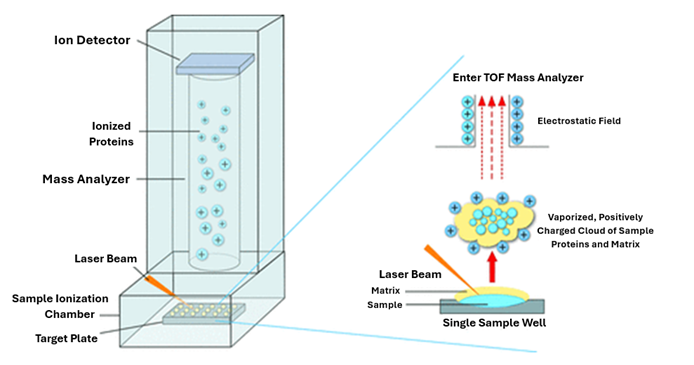
Only a small amount of test material is required to perform identification – in practice, it is a single bacterial colony grown on a solid medium. First, the test material is transferred to a plate, and a special matrix solution is applied. The purpose of this solution is to strongly absorb the light energy of the laser, which leads to the release (desorption) of analyte molecules. Once prepared and dried, the sample is placed in the analyzer chamber, where the laser’s action causes desorption and ionization of matrix molecules and bacterial proteins.
Next, the released particles are subjected to acceleration, and then measured and recorded by the detector. Low mass particles are the first to reach the detector, followed by increasingly larger particles, resulting in the formation of a mass spectrum – a unique “fingerprint” specific to each species of the studied microorganisms. This spectrum is automatically analyzed by comparing it with a database containing spectra characteristic of specific microbial species (Figure 1 and Figure 2). An outstanding advantage of this system is its ability to read as many as 96 isolates in just 1,5 hours.4
When comparing the mass spectrum of the studied microorganism with a reference spectrum, the so‑called identification confidence level is determined. Depending on how high this level is, the organism can be assigned to a family, genus or species. Various algorithms are used to compare bases. The MALDI Biotyper system (Bruker) recognizes patterns and, on the basis of presence or absence of peaks in the bases, determines the so‑called score index, which can range from 0 to 3,000. According to the manufacturer’s criteria, a score of ≥2,000 translates to identification at the species level, a score of 1,700 – 1,999 at the genus level, and a score of ≤1,700 means no identification. Search through the database for MALDI VITEK mass spectrometry system is performed with the use of 1 of the 2 algorithms. The first one is a pattern matching process otherwise known as the reference spectra approach. In the second approach, mass spectra are grouped together as “SuperSpectra,” which represent the spectra of conservative mass signals from multiple individual isolates of a specific group of microorganisms. Confidence values for the output range from 0% to 100%.6
Applications of the MALDI mass spectrometry method in microbiological diagnostics
Environmental bacteriology
Due to the huge diversity of microorganisms in samples isolated from the environment, species identification based on biochemical characteristics is usually not feasible. Various studies have shown that MALDI-TOF MS is an effective tool for identification and characterization of microorganisms from different ecosystems.8 This method has method has successfully identified bacteria isolated from sewage sludge, soil, water and even marine sponges or bioaerosol particles. Additionally, MALDI has been evaluated for rapid identification and differentiation of coliform bacteria in surface water. The method also allows to identify microorganisms potentially pathogenic to humans in drinking water, leading to conclusion that this technique can be successfully applied to the detection of microbial infections in water.7
Food-borne bacteria
Reliable and rapid identification of microorganisms in food products is crucial for ensuring their quality. Among other things, food spoilage causing bacteria, foodborne pathogens, probiotics and starter cultures need to be accurately detected. It is the only way to prevent fermentation errors, uncover potential pathways for transmission of contaminants and improve hygienic conditions in production facilities. Timing is a key factor in food processing as contamination in the production process can go unnoticed for several days, negatively impacting the product’s safety. Species identification of bacteria isolated from food using MALDI-TOF MS techniques has so far focused on food pathogens such as Listeria spp., Campylobacter spp., Vibrio sp. and Salmonella spp. Only a few studies show application of these methods in identification of microorganisms from food and beverage sources in which they have been used, such as the surveillance of probiotics in yogurt or characterization and identification of amine-producing bacteria.8
Clinical bacteriology
Conventional bacteriological diagnosis of body fluids is carried out using metabolic and biochemical profiling of bacteria. This diagnosis requires 24 – 48 hours to identify the causative ganglion. In the meantime, the patient is given antibiotics, which may not be appropriate in treating the ongoing infection. This is why it has become so important to look for fast, accurate and cost‑effective methods to identify microorganisms in clinical trials. A number of researchers have demonstrated that MALDI-TOF MS can be successfully used for early identification of bacteria in cerebrospinal fluid, blood cultures, urinary tract infections and respiratory tract infections, allowing rapid application of appropriate antimicrobial therapy.9
Detection of Antibiotic Resistance
Due to the increasing prevalence of multidrug-resistant strains, especially in hospital settings, there is an urgent need to implement rapid methods for determining antimicrobial susceptibility of microorganisms. Several studies have verified the feasibility of using the MALDI-TOF MS method to detect antibiotic resistance in bacteria isolated from bloodstream infections and in pathogenic fungi.10 The main principle of this procedure is to detect the so-called “resistance peak pattern,” i.e., characteristic peak differences in the mass spectra of resistant and antibiotic-sensitive microorganisms using the classical strain typing methodology.11
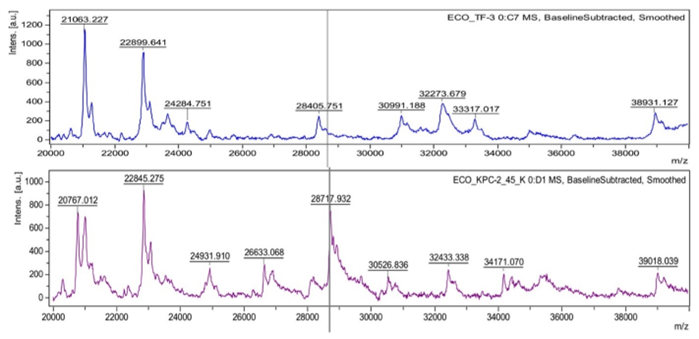
Bacteria isolated from pharmaceutical clean rooms
During the manufacturing of sterile products, it is extremely important to adhere to the aseptic requirements stipulated in the good manufacturing practice (GMP) guidelines. Rigorous and comprehensive procedures must be implemented in order to obtain a safe product. One of the most important requirements that must be met is the maintenance of adequate cleanliness of production rooms. Clean rooms are controlled environments that require an effective monitoring program and therefore a rapid and trustworthy method for detecting undesirable microbiological events is needed so that the specific corrective actions can be introduced. The primary purpose of monitoring such rooms is to measure the number of particles as well as the number and diversity of microorganisms, and compare the obtained values with the accepted limits. Knowledge of the natural microflora of the manufacturing environment is important because it can provide useful information for the investigation of contamination source, especially when action limits are exceeded. In addition, the identification of this microflora is essential for selecting microorganisms for validation processes or evaluating the effectiveness of disinfectants.
Conventional methods available to microbiology laboratories are mainly based on culture methods, which is often a time-consuming and costly process. Therefore, pharmaceutical companies focus on developing new, more efficient techniques to accelerate the analyses and improve their reliability. MALDI-TOF MS method fits perfectly into the sought standards. For example, a study conducted by Laíse de Oliveira Andrade et al. showed excellent identification performance for microorganisms isolated from clean rooms with the MALDI-TOF MS technique compared to the conventional method (biochemical tests) in terms of correct species identification (88.89% and 38.89%, respectively). One barrier to the wider implementation of this method in pharmaceutical companies is the smaller library of spectra for environmental strains compared to clinical isolates. As the variability of environmental microorganisms is very high and interferes with method’s accuracy, further improvements in libraries and sample preparation techniques are clearly needed. Nevertheless, this fast, inexpensive and simple method can be successfully implemented in conventional quality control laboratory settings, provided that close attention is paid to improving the databases.13
Mycology and virology
Conventional methods for identifying fungal species are based on their morphological, immunological and biochemical properties. These methods typically require 2 to 5 days and often combine several phenotypic techniques to produce a definitive result. MALDI-TOF MS is also used for this purpose, but the identification process is slower than for bacteria due to greater biological complexity and coexistence of different phenotypes within the same organism. In order to obtain reproducible results, parameters must be carefully standardized and additional treatment may be required in order to break strong fungi cell walls.
The use of MALDI-TOF in virology is at less advanced stage than in either bacteriology or mycology, which may be due to, among other things, relatively low protein content of viral particles or their higher molecular weight. Nevertheless, many researchers have demonstrated the usefulness of MALDI method in identifying certain viruses in clinical samples including influenza, HPV and hepatitis viruses. The detection limit and sensitivity were reported to be high and comparable to the reference methods.9
Conclusions
In conclusion, identification of microorganisms based on MALDI-TOF MS method has found application in numerous laboratories and industries. This technique is fast, precise and relatively inexpensive. Thanks to its multi-level functionality, it can successfully serve as an alternative to more traditional microbial identification tools. The ability of MALDI-TOF mass spectrometry to accurately and rapidly detect potential threats enables efficient responses and helps maintain a high level of microbiological safety.
Prepared by:
Karolina Kromkowska
References
- Israr MZ, Bernieh D, Salzano A, Cassambai S, Yazaki Y, Suzuki T. Matrix-assisted laser desorption ionisation (MALDI) mass spectrometry (MS): basics and clinical applications. Clin Chem Lab Med. 2020; 58(6): 883-896.
- Hrabák J, Chudácková E, Walková R. Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry for detection of antibiotic resistance mechanisms: from research to routine diagnosis. Clin Microbiol Rev. 2013; 26(1): 103-14.
- Lay JO Jr. MALDI-TOF mass spectrometry of bacteria. Mass Spectrom Rev. 2001; 20(4): 172-94.
- Cieślik J, Wróblewska M. MALDI TOF MS – nowe możliwości w rutynowej diagnostyce mikrobiologicznej. Diagnostyka Laboratoryjna. 2018; 54(2): 99-104.
- Krásný L, Hynek R, Hochel I. Identification of bacteria using mass spectrometry techniques, Int. J. Mass Spectrom. 2013; 353: 67-79.
- Patel R. MALDI-TOF MS for the diagnosis of infectious diseases. Clin Chem. 2015; 61(1): 100-11.
- Ashfaq MY, Da’na DA, Al-Ghouti MA. Application of MALDI-TOF MS for identification of environmental bacteria: A review. J Environ Manage. 2022; 305: 114359.
- Pavlovic M, Huber I, Konrad R, Busch U. Application of MALDI-TOF MS for the Identification of Food Borne Bacteria. Open Microbiol J. 2013; 7: 135-41.
- Singhal N, Kumar M, Kanaujia PK, Virdi JS. MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front Microbiol. 2015; 6: 791.
- Florio W, Baldeschi L, Rizzato C, Tavanti A, Ghelardi E, Lupetti A. Detection of Antibiotic-Resistance by MALDI-TOF Mass Spectrometry: An Expanding Area. Front Cell Infect Microbiol. 2020; 10: 572909.
- Vrioni G, Tsiamis C, Oikonomidis G, Theodoridou K, Kapsimali V, Tsakris A. MALDI-TOF mass spectrometry technology for detecting biomarkers of antimicrobial resistance: current achievements and future perspectives. Ann Transl Med. 2018; 6(12): 240.
- Yoon EJ, Jeong SH. MALDI-TOF mass spectrometry Technology as a Tool for the Rapid Diagnosis of Antimicrobial Resistance in Bacteria. Antibiotics (Basel). 2021; 10(8): 982.
- Andrade LO, Awasthi R, Dua K, de Jesus Andreoli Pinto T. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of bacteria isolated from pharmaceutical clean rooms. Interv Med Appl Sci. 2018; 10(1): 45-53.
Related resources

Qualitative analysis of Host Cell Proteins using mass spectrometry
Analytics, Drug development, Drug product, Drug substance

Does Your CDMO Have An Analytical Edge?
Analytics, Biosimilars, Mabion

Innovative biologics – expected drug approvals in 2025
Antibody-drug conjugates, Biologics, Bispecific antibody, Clinical trials, EMA, FDA, Monoclonal antibody, Vaccines